How to deal with huge systems?
- Fixing custom ICs.
- Fixing by secondary structure.
- Fixing ICs randomly.
imodfit ccmvfit_fitted.pdb ccmv_swln_10A3.ccp4 10 0 -n 0.99999 --prob plain -i 300000 -r 1 -t -o ccmvfit2
How to avoid Secondary Structure distortions?
To avoid Secondary Structure distortions, the SS elements can be fixed. This may result specially useful dealing with huge systems and/or experimental maps.
The easiest procedure is fixing both the IC of α-helices and β-strands. To this end add the "-S HE" option. This way the unique mobile ICs will be the coils dihedral angles and inter-segment rotations/translations:
The "HE" characters introduced after "-S" parameter are the single character identifier for α-helices and "E" for β-strands. In addition, you may fix any combination of SS with as many SS identifiers as you need; for example, "−S EC" will fix beta and coil SS ICs.
By default, the SS is computed internally existing three SS types available: "H", "E" and "C" (coil); but any user defined SS can be provided using the −−ss option, as long as the SS identifiers are single characters. For example, you can use DSSP to compute SS ( 1aon.dssp ) and with the aid of this simple perl script you can convert it into our SS file format (1aon.ss).
An increase in the number of iterations (−i 30000) was needed in order to ensure convergence.
In the screen output you can check both the dimensionality ("Mobile Internal Coordinates") and diagonalization time ("NMA_time") reduction respecting to the basic command (tutorial section):
imodfit> imodfit> Welcome to iMODFIT v1.28 imodfit> imodfit> Model PDB file: 1aon.pdb molinf> Protein 1 chain 1 segment 1 residues: 524 atoms: 3847 molinf> SUMMARY: molinf> Number of Molecules ... 1 molinf> Number of Chain ....... 1 molinf> Number of Segments .... 1 molinf> Number of Groups ...... 524 molinf> Number of Atoms ....... 3847 molinf> imodfit> Coarse-Graining model: Full-Atom (no coarse-graining) imodfit> Selected model number of residues: 524 imodfit> Selected model number of (pseudo)atoms: 3847 imodfit> Target Map file: 1oel.ccp4 imodfit> Best filtration method: 2 FT(x10)=0.100s Kernel(x10)=0.080s imodfit> Number of Inter-segment coords: 0 (Rot+Trans) imodfit> Number of Internal Coordinates: 1033 (Hessian rank) imodfit> Input CG-model Fixed Internal Coordinates: 668 imodfit> Input CG-model Mobile Internal Coordinates (size) = 365 imodfit> Range of used modes: 1-73 (20.0%) imodfit> Number of excited/selected modes: 1(nex) imodfit> imodfit> Iter score Corr. NMA NMA_time imodfit> 0 0.336409 0.663591 0 0.75 sec imodfit> 199 0.326737 0.673263 1 0.76 sec imodfit> 384 0.286446 0.713554 2 0.68 sec ................................................. imodfit> 8850 0.032497 0.967503 17 0.71 sec imodfit> 30000 0.024034 0.975966 END imodfit> imodfit> Movie file: imodfitHE_movie.pdb imodfit> Final Model: imodfitHE_fitted.pdb imodfit> Score file: imodfitHE_score.txt imodfit> Log file: imodfitHE.log imodfit> imodfit> Success! Time elapsed 00h. 09' 59'' imodfit> Bye!
The fitting result ( ![]() imodfitHE_fitted.pdb) is only 1.99Å Cα RMSD from the target structure , and the final correlation was high: 0.976. The quality of fitness and the excellent secondary structure maintenance can be appreciated in the flash movie below. In addition you can play interactively the trajectory movie in
imodfitHE_fitted.pdb) is only 1.99Å Cα RMSD from the target structure , and the final correlation was high: 0.976. The quality of fitness and the excellent secondary structure maintenance can be appreciated in the flash movie below. In addition you can play interactively the trajectory movie in ![]() Jmol.
Jmol.
How to fix domains?
Any domain/s in our macromolecule can be fixed using the −f option. This way both dimensionality will be reduced and undesired distortions will be avoided. For illustrative purposes we will fix two GroEL domains.
GroEL is composed of three domains: apical (top red), intermediate (cyan), and equatorial (bottom red). The apical domain interacts with folding intermediates and GroES, the equatorial domain hydrolyzes ATP, and the intermediate domain is flexible and connects these apical and equatorial domains.
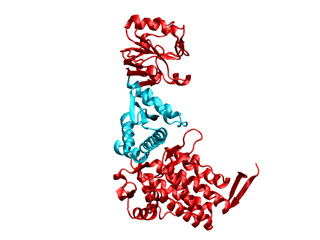 |
To obtain a successful fitting the structure should be maintained as flexibile as possible, i.e. if we fix very much the structure will be very rigid and the target conformation will not be addressed. To this end the apical and equatorial domains will be fixed while keeping the intermediate domain fully flexible. In the mask file imodfitDOM.fix only the ICs belonging to regions of the intermediate domain are keept mobile: 136-193 and 331-409 (indexed from 1 to 524). Further information about the mask file generation is provided in iMod tips section (to obtain a dummy mask (fully mobile) use the command: imode 1aon.pdb --save_fixfile -o imodfitDOM)
The output is the following:
imodfit> imodfit> Welcome to iMODFIT v1.28 imodfit> imodfit> Model PDB file: 1aon.pdb molinf> Protein 1 chain 1 segment 1 residues: 524 atoms: 3847 molinf> SUMMARY: molinf> Number of Molecules ... 1 molinf> Number of Chain ....... 1 molinf> Number of Segments .... 1 molinf> Number of Groups ...... 524 molinf> Number of Atoms ....... 3847 molinf> imodfit> Coarse-Graining model: Full-Atom (no coarse-graining) imodfit> Selected model number of residues: 524 imodfit> Selected model number of (pseudo)atoms: 3847 imodfit> Target Map file: 1oel.ccp4 imodfit> Best filtration method: 2 FT(x10)=0.090s Kernel(x10)=0.080s imodfit> Number of Inter-segment coords: 0 (Rot+Trans) imodfit> Number of Internal Coordinates: 1033 (Hessian rank) imodfit> Input CG-model Fixed Internal Coordinates: 760 imodfit> Input CG-model Mobile Internal Coordinates (size) = 273 imodfit> Range of used modes: 1-54 (19.8%) imodfit> Number of excited/selected modes: 1(nex) imodfit> imodfit> Iter score Corr. NMA NMA_time imodfit> 0 0.336409 0.663591 0 0.73 sec imodfit> 199 0.324457 0.675543 1 0.50 sec imodfit> 379 0.285427 0.714573 2 0.49 sec ................................................. imodfit> 12857 0.080700 0.919300 14 0.50 sec imodfit> 50000 0.064124 0.935876 END imodfit> imodfit> Movie file: imodfitDOM_movie.pdb imodfit> Final Model: imodfitDOM_fitted.pdb imodfit> Score file: imodfitDOM_score.txt imodfit> Log file: imodfitDOM.log imodfit> imodfit> Success! Time elapsed 00h. 16' 18'' imodfit> Bye!
The solution is only 3.77 Å Cα RMSD from the target structure used to simulate the map. This is a good result for a structure with 75% of its ICs fixed. Note that, to obtain a good result with such a fixed structure, the number of iterations was increased (-i 50000). If you increase the number of iterations even more you will reach lower RMSD.
You can play interactively the trajectory movie in ![]() Jmol, check the fitted structure
Jmol, check the fitted structure ![]() imodfitDOM_fitted.pdb and watch the flash movie below. For illustrative purposes the fixed domains were colored in red.
imodfitDOM_fitted.pdb and watch the flash movie below. For illustrative purposes the fixed domains were colored in red.