-
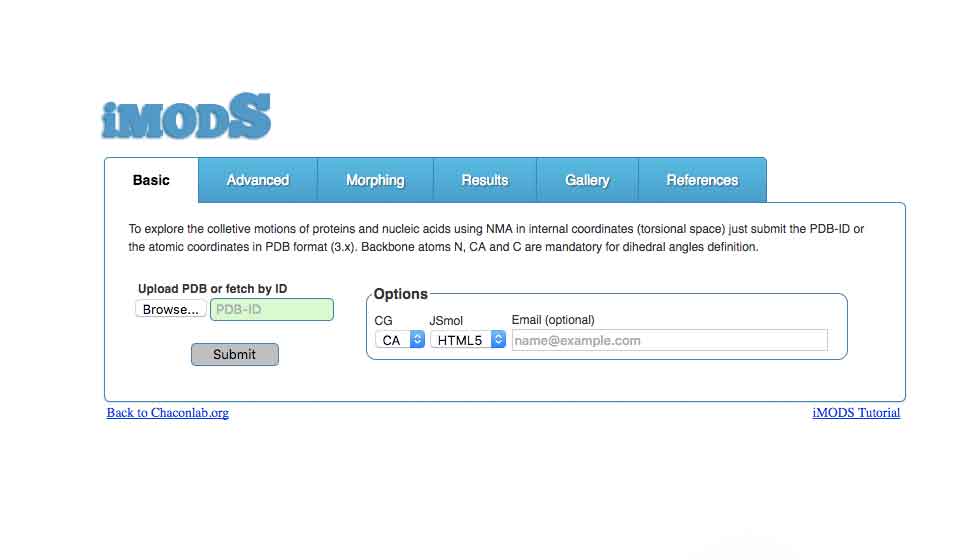
Collective motions prediction
imods.chaconlab.org
To explore the collective motions of proteins and nucleic acids using NMA in internal coordinates (torsional space) just submit the PDB-ID or the atomic coordinates in PDB format (3.x).
-
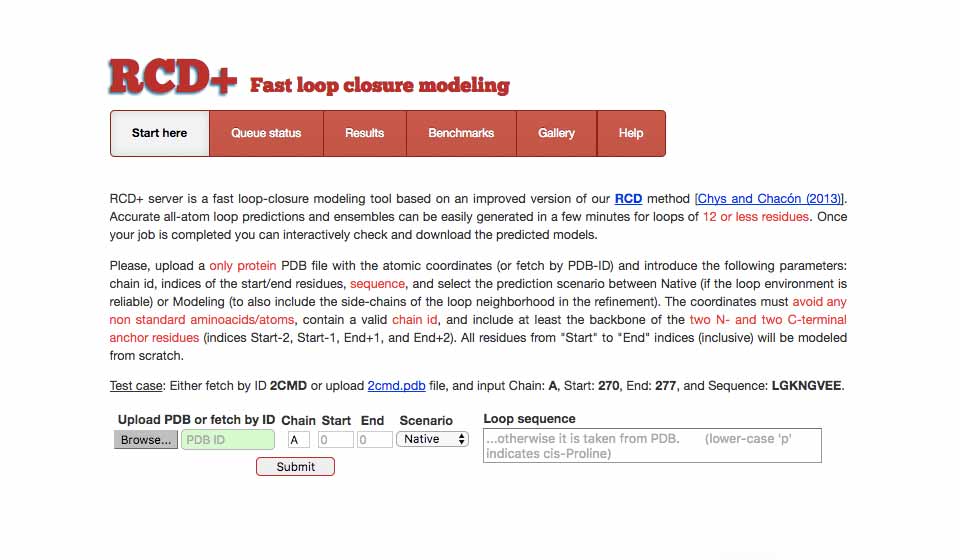
RCD+ Ab initio loop modeling
rcd.chaconlab.org
RCD+ server is a fast loop-closure modeling tool based on an improved version of our RCD method. Accurate all-atom loop predictions and ensembles can be easily generated in a few minutes.
-
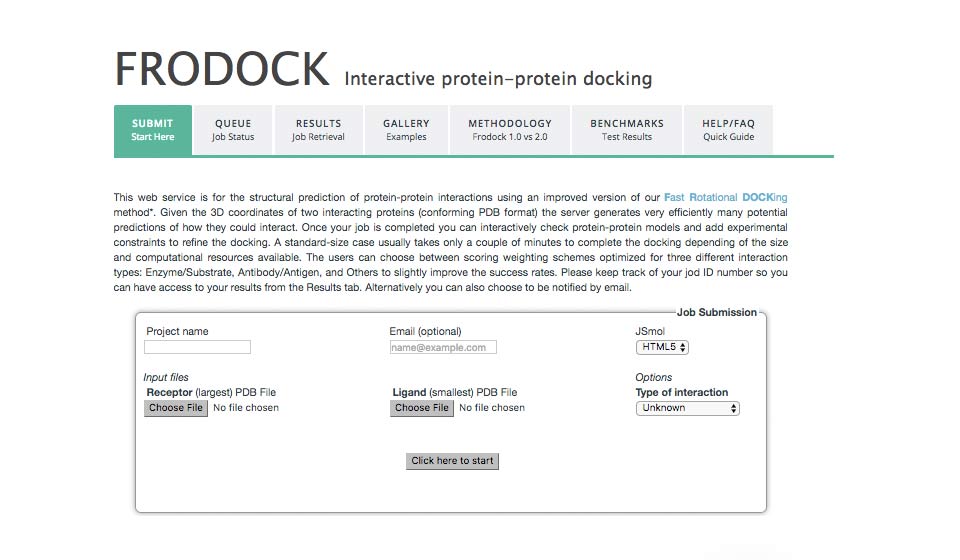
Frodock Protein-protein docking
frodock.chaconlab.org
This web service is for the structural prediction of protein-protein interactions using an improved version of our Fast Rotational DOCKing method*. Given the 3D coordinates of two interacting proteins (conforming PDB format) the server generates very efficiently many potential predictions of how they could interact.