How to fit two volumes?
ADP_EM not only provides a way to fit atomic structures inside a low-resolution maps. It allows also the rigid-body fitting of two tridimensiional maps. This fearture provide a way to compare different experimental maps of the same molecule or to localize the low-resolution information of a molecule inside the low map of a macromolecular complex.
The process to fit a tridimensional map into another map is quite similar to the case when a atomic structure is fitted. The only aspects that must be changed in the invocation of the command are:
- Instead of a PDB file, the second argument must be a CCP4/SIT format file containing the map to be fitted.
- The fith argument, instead of defining the resolution, will set the density cutoff for the map to be fitted. Notice that in thtis case the resolution value have no sense since we have not a pdb to be filtered.
Example:
In this case we have simulated a 14.9 resolution map (right) from a single chain of 60 KDA CHAPERONIN (2C7E.pdb):
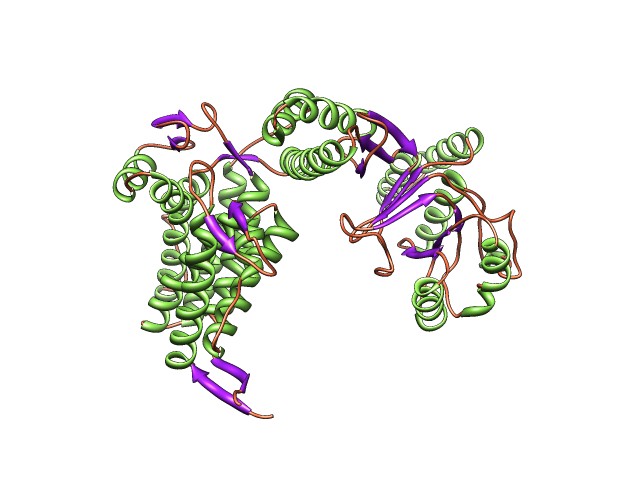 |
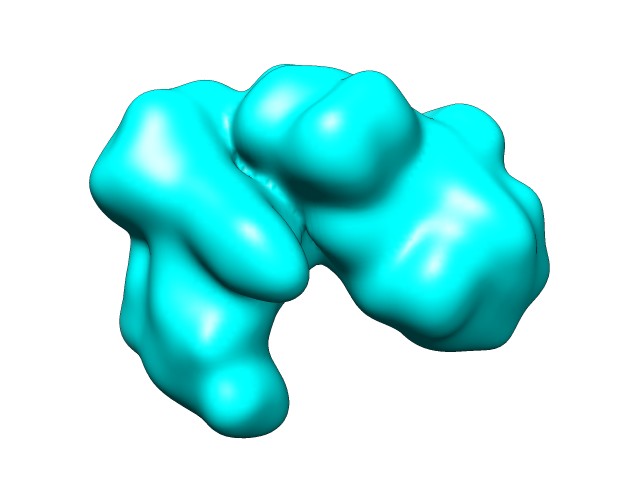 |
This map will be fitted in the experimental map emd1047.ccp4 (download from EMD, removed discontinuos density using voledit tool of SITUS package. Resolution 14.9 and floodfill at 0.060):
 |
To fit the chain map, adp_em must be invoqued in the next way:
This will return:
adp_em>
adp_em> ADP_EM
adp_em>
adp_em> EM map
adp_em> Laplacian FT Filtering
adp_em> Search Mode: Mask
adp_em> Save 100 best fits
adp_em> Sampling-> Rot: 11.250 (bw=16) Trans: 2.000
adp_em>
adp_em>
adp_em> Step 1: Processing EM-map & EM-map...
adp_em>
adp_em> 3DMAP ../emd1047.ccp4
adp_em> Resolution 16.000000 cutoff 0.060000
adp_em> Initial dimensions 237x237x237
adp_em> Grid Size 1.000x1.000x1.000
adp_em> Mass center 74.201x74.203x82.158
adp_em> Radii E 15 99 I 10 76 Z 0 0
adp_em> Radii E 18 102 I 7 32 Z 0 0
adp_em> Final dimensions 152x153x161 voxels
adp_em>
adp_em> 3DMAP chainH_14.9.sit
adp_em> DIM: Low pass 72x68x86
adp_em> Radii E 5 47 I 0 0 Z 6 48
adp_em> Final dimensions 74x70x88
adp_em>
adp_em> Step 2: Search...
adp_em>
adp_em> Trans limits min 7.000 max: 97.000 init 7.000
adp_em> Explored 188156 (mask 1505750 not 285856): time -772.12 sec
adp_em> Max peaks 100 (allowed 100)
adp_em> Peaks found 13 (from 100 peaks) time: -745.41 sec
adp_em>
adp_em> Total searching time 3571.000000 sec
adp_em>
adp_em>
adp_em> Step 3: Saving Solutions...
adp_em>
adp_em> ---------------------------------------------------------------------
adp_em> Psi Theta Phi X Y Z Corr
adp_em> ---------------------------------------------------------------------
adp_vol_em> 1 231.56 -0.00 74.24 122.00 62.00 116.00 0.280
adp_vol_em> 2 318.03 -0.00 47.98 114.00 104.00 118.00 0.278
adp_vol_em> 3 271.15 -0.00 248.68 28.00 66.00 118.00 0.277
adp_vol_em> 4 236.86 -0.00 23.14 94.00 30.00 116.00 0.276
adp_vol_em> 5 301.21 -0.00 109.98 78.00 124.00 116.00 0.273
adp_vol_em> 6 226.15 -0.00 237.29 40.00 106.00 116.00 0.270
adp_vol_em> 7 345.04 -0.00 221.22 52.00 32.00 118.00 0.266
adp_vol_em> 8 285.06 173.71 266.11 104.00 38.00 42.00 0.252
adp_vol_em> 9 318.54 174.10 198.85 32.00 54.00 44.00 0.247
adp_vol_em> 10 320.77 174.30 106.70 64.00 122.00 44.00 0.245
adp_vol_em> 11 314.32 180.00 146.96 32.00 96.00 42.00 0.244
adp_vol_em> 12 312.89 173.18 249.85 64.00 28.00 44.00 0.242
adp_vol_em> 13 309.71 171.51 353.51 122.00 74.00 44.00 0.238
adp_em> ----------------------------------------------------------------------
adp_em>
adp_em> Saved in adpEM[1-13].sit files
adp_em> Total Time 3617.000000 sec
adp_em>
adp_em>
Here, in a similar fashion to the atomic structure fitting case, the best rotations and translations showed in the log are applied to have the corresponding map solution files. Here can be observed the first solution from different points of view:
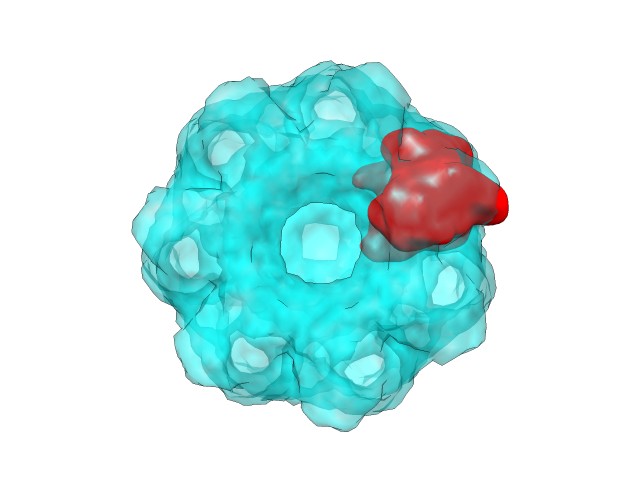 |
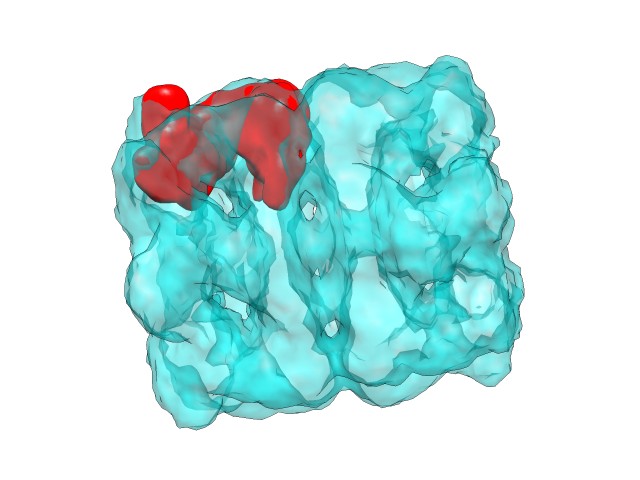 |
And here the reconstruction of one of the rings from the combination of the first solutions returned:
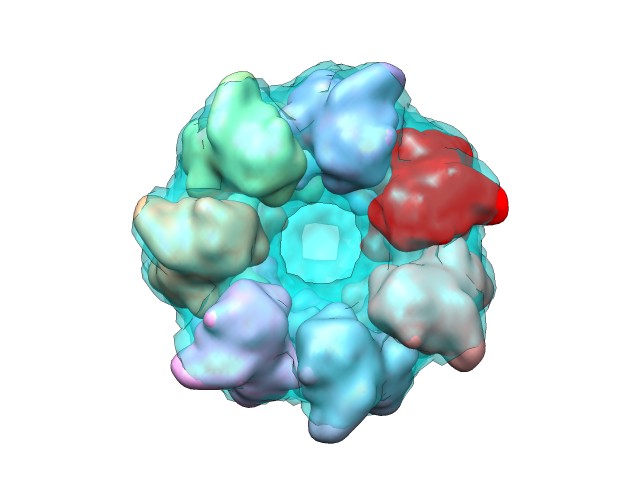 |
How to perform a multiple comparative fitting?
- The second argument (after the input map) must be a text file containing the paths and names of the PDB files with all the atomic structures to fit. There must contain a PDB name per line as:
../1dxt/1dxt.pdb
../1dxt/1hbg.pdb
../1dxt/model100.pdb
../1dxt/model101.pdb
../1dxt/model102.pdb
../1dxt/model103.pdb
- Set the flag -m. Example:
ADPEM will return a list of results where the last column shows corresponding fitted PDB:
1 259.12 -0.00 101.62 30.00 32.00 34.00 0.986 ../1dxt/1dxt.pdb
1 229.33 148.66 219.85 30.00 30.00 32.00 0.938 ../1dxt/1hbg.pdb
1 184.21 161.12 164.73 32.00 32.00 32.00 0.929 ../1dxt/model100.pdb
1 149.93 159.74 141.41 30.00 32.00 34.00 0.901 ../1dxt/model101.pdb
1 132.01 136.90 104.85 32.00 32.00 32.00 0.945 ../1dxt/model102.pdb
1 251.51 156.19 231.27 30.00 32.00 34.00 0.905 ../1dxt/model103.pdb
1 153.28 146.61 140.50 32.00 32.00 32.00 0.947 ../1dxt/model104.pdb