This is a practical guide for researchers who want to perform protein-protein docking using the FRODOCK docking tool. Please, follow the next working examples:
- Step by step simple protein-protein docking case
- Using unbound ligand pdb in docking
- MPI parallel docking
In the install section you can find the links to the necesary files to complete the tutorial. Please uncompress and untar the corresponding archive and go to the examples directory.
1) Step by step easy protein protein docking
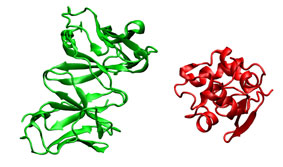
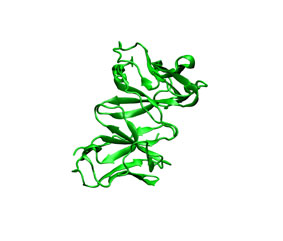
To illustrate the procedure it is performed a HyHel-5/lysozyme bound-bound docking case. The receptor (3hfl_fv.pdb, green) and the ligand (3hfl_ly_ref.pdb, red on the right) coordinates were extracted from 3hfl pdb entry of the Protein Data Bank. The ligand molecule was rotated randomly (3hfl_ly2.pdb, red on the left)to avoid pre-aligment situations.
 |
FRODOCK => | 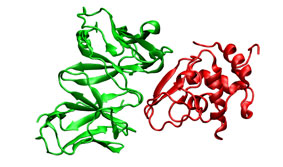 |
The docking process will be carried out in three consecutive steps:
--------------- STEP 1. Check PDB input files ---------------
First, the input coordinates of both ligand and receptor should conform to PDB format. Be aware of missing atoms, alternative conformations, bad place atoms and a long etc. that can eventually jeopardize your results. Use your favorite PDB checker to anticipate and fix any PDB error. We typically employ Molprobity (online), pdb2pqr (e.g. pdb2pqr file.pdb --ff=CHARMM ), profix from Jackal package or a combination of them.
--------------- STEP 2. Potential map generation ---------------
All necessary potential maps must be pre-computed using FRODOCKGRID. Although vdw and electrostatics maps could be computed on the fly during the docking search, it is recommendable to create the maps beforehand in order to visualize them and check that they are consistent with the original structure. The precomputation of desolvation potential maps for receptor and ligand is always required.
Creation of receptor vdw potential map:
Name convention: iupac
Force Field: CHARMM
Grid voxel: 1.0
With hydrogens: no
Option: vdw potential. Probe radius: 2.0
Creation of the receptor electrostatic potential map:
Name convention: iupac
Force Field: CHARMM
Grid voxel: 1.0
With hydrogens: no
Option: Electrostatic potential.
Creation of the receptor desolvation potential map:
Name convention: iupac
Force Field: CHARMM
Grid voxel: 1.0
With hydrogens: no
Option: Desolvation potential.
Radius -> Water: 1.400000 - Generic atom probe: 1.950000
Area: 10102.59703 Spheres num: 1661
ASA pdb file 3hfl_fv_ASA.pdb created (ASA introduced in Occupancy column)
Computing Map..
ASA computations: 620259 ASA computations skipped: 344332
Also notice that, together with the desolvation potential map 3hfl_fv_desol.ccp4, this command creates a version of 3hfl_fv.pdb called 3hfl_fv_ASA.pdb that stores atom ASA values in the occupancy column. This pdb file MUST be used when desolvation term is employed in the docking process.
You can check the appearance of the generated receptor potential maps with a viewer that supports ccp4 or situs. As an example, next it is shown from left to right and from top to bottom: the receptor structure, van der Waals, electrostatic, and desolvation maps. All of them were represented in the same pose using blue color for negative and red for positive potentials. For better display arbitrary visualization thresholds were used.
 |
 |
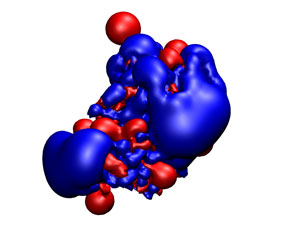 |
 |
Creation of the ligand desolvation potential map:
Name convention: iupac
Force Field: CHARMM
Grid voxel: 1.0
With hydrogens: no
Option: Desolvation potential.
Radius -> Water: 1.400000 - Generic atom probe: 1.950000
Area: 6339.83754 Spheres num: 1001
ASA pdb file 3hfl_ly2_ASA.pdb created (ASA introduced in Occupancy column)
Computing Map..
ASA computations: 385533 ASA computations skipped: 210271
To consider desolvation in the docking process desolvation potentials for both receptor and ligand must be provided. As the receptor desolvation case, the command creates also a pdb file 3hfl_ly2_ASA.pdb with ASA values that MUST employed in the docking process.
As it previous case, you can check the appearance of the generated ligand desolvation potential map (see below) The same convention as above was employed.
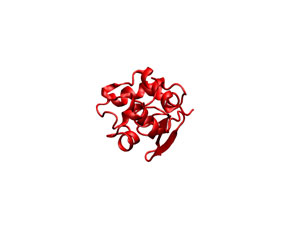 |
 |
--------------- STEP 3. Perform the exhaustive docking search ---------------
Once the potential maps were generated, you are ready to perform the docking. To this end, use the the following command performs to include all the potentials (van der Waals, Electrostatic and desolvation):
dock.dat is a binary file which stores all the docking solutions (ligand orientations and positions). Here it is the screen output:
Reading receptor PDB
* Receptor accessibility read from receptor PDB.
Checking ASA in Occupancy column...
- Ok. Occupancy column seems to show ASA values
* Creating Accessibility map from receptor PDB...
* Accessibility map created from receptor PDB
* OK
Receptor VDW map information:
* Receptor VDW map read from file
* Map origin: -33.141129 -39.668289 -12.076756
* Map size: 89 81 75
Receptor electrostatic map information:
* Receptor electrostatic map read from file
* Map modification. Threshold applied: 10.000000
* Map origin: -33.141129 -39.668289 -12.076756
* Map size: 89 81 75
Receptor desolvation map information:
* Receptor electrostatic map read from file
* Map origin: -24.141129 -30.668287 -3.076756
* Map size: 71 63 57
* Extra potential receptor maps
*Number of read potentials: 0
Reading ligand pdb
* Ligand center: 39.044571 33.064610 11.924802
* Ligand accesilibility read from Ligand PDB.
Checking ASA in Occupancy column...
- Ok. Occupancy column seems to show ASA values
* Ligand desolvation map read from file
Spherical radius
* Receptor vdw radius in amstrongs: 71.000000 (71 spherical layers)
* Ligand radius in amstrongs: 25.011627 (26 spherical layers)
* Ligand electrostatic radius in amstrongs: 25.011627 (26 spherical layers)
* Ligand accesibility radius in amstrongs: 25.011627 (26 spherical layers)
* Ligand desolvation radius in amstrongs: 30.011627 (31 spherical layers)
* Maximum ligand desolvation radius in amstrongs: 30.011627 (31 spherical layers)
Precomputation of spherical representation of ligand
Precomputation Fixed time: 42.360000 s
Determination of translational points to explore
* Mask created
* Number of points to explore: 20356
Translational search init...
* Translational search finished
* Number of solutions: 70801
Note for advanced users: The van der Waals potential term cannot be disabled; if it is not given as input, it will be automatically computed and used. In order to avoid the use of the electrostatic potential −−E option must be set to 0.0. To avoid the use of desolvation and ASA projection maps, just don't introduce them.
--------------- STEP 4. Clustering and visualization of predictions ---------------
Once the docking process has been performed it is possible to sort and cluster the solutions by their RMSD values. To this end:
This way a new solution file containing only 100 solutions is created. Each of these solutions represents the best element of the first 100 clusters created. The RMSD default value used in the clusterization is 5Å.
Sort process...
Maximum correlation: 1359.015259
Clustering process...
To visualize the 10th first solutions:
1 86.00 158.32 201.22 19.36 6.83 55.42 1359.015259
2 244.84 55.47 123.03 13.36 2.83 51.42 1340.471680
3 194.98 99.53 265.77 -6.64 14.83 9.42 1331.359131
4 314.93 81.77 144.80 5.36 2.83 51.42 1314.301514
5 307.26 142.28 57.47 7.36 6.83 51.42 1297.874390
6 350.57 90.12 270.98 -0.64 20.83 15.42 1283.359741
7 243.61 82.73 26.85 11.36 2.83 57.42 1268.607544
8 312.64 125.93 92.45 5.36 10.83 49.42 1267.174683
9 273.09 101.75 35.90 3.36 4.83 55.42 1237.752808
10 149.5 105.01 199.2 -0.64 18.83 11.42 1225.663574
Eeach line has the format:
rank Euler1 Euler2 Euler3 posX posY PosZ correlation
where rank indicates the position of the solution, {Euler1,Euler2,Euler3} are the Euler angle rotation (in ZYZ convention) and {posX ,posY ,PosZ} is the X,Y,Z localization respect the center of mass of the ligand pdb (only C-alpha are used to center). Finally, the correlation is the absolute energy score obtained.
Coordinate generation of the predicted solutions:
1 86.00 158.32 201.22 19.36 6.83 55.42 1359.01 => 3hfl_ly2_1.pdb
2 244.84 55.47 123.03 13.36 2.83 51.42 1340.47 => 3hfl_ly2_2.pdb
3 194.98 99.53 265.77 -6.64 14.83 9.42 1331.35 => 3hfl_ly2_3.pdb
4 314.93 81.77 144.80 5.36 2.83 51.42 1314.30 => 3hfl_ly2_4.pdb
5 307.26 142.28 57.47 7.36 6.83 51.42 1297.87 => 3hfl_ly2_5.pdb
with this command the ligand pdb is rotated and translated using to generate the first 5 predictions. For each prediction a pdb file is created. In this case, the first prediction (3hfl_ly2_3.pdb) shows to be very similar to the real solution (3hfl_ly2_ref.pdb).
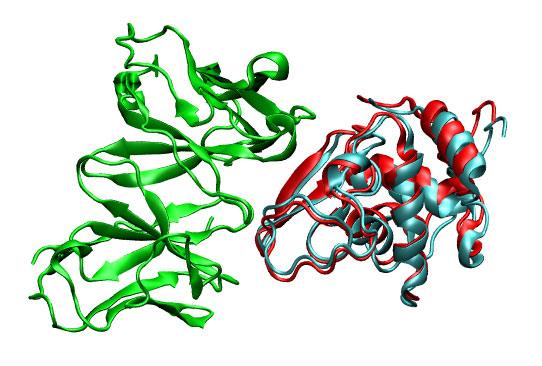 |
2) Using unbound ligand pdb in docking
In this case, instead of using the ligand bound structure obtained from the complex, it is employed an unbound ligand structure (PDB entry 1lza). Being an unbound structure, it presents conformational changes that makes more difficult predict the correct position.
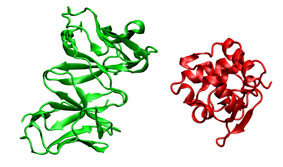 |
FRODOCK ==> |  |
The steps to follow are very similar to the ones performed in the previous tutorial:
--------------- STEP 2. Potential map generation ---------------
The receptor potential maps were already created in the previous section, thus only the desolvation term must be pre-computed:
Creation of the unbound ligand desolvation potential map:
Name convention: iupac
Force Field: CHARMM
Grid voxel: 1.0
With hydrogens: no
Option: Desolvation potential.
Radius -> Water: 1.400000 - Generic atom probe: 1.950000
Area: 6546.14710 Spheres num: 1001
ASA pdb file 1lza_ly2_ASA.pdb created (ASA introduced in Occupancy column)
Computing Map..
ASA computations: 388332 ASA computations skipped: 209303
--------------- STEP 3. Perform the exhaustive docking search ---------------
Reading receptor PDB
* Receptor accessibility read from receptor PDB.
Checking ASA in Occupancy column...
- Ok. Occupancy column seems to show ASA values
* Creating Accessibility map from receptor PDB...
* Accessibility map created from receptor PDB
* OK
Receptor VDW map information:
* Receptor VDW map read from file
* Map origin: -33.141129 -39.668289 -12.076756
* Map size: 89 81 75
Receptor electrostatic map information:
* Receptor electrostatic map read from file
* Map modification. Threshold applied: 10.000000
* Map origin: -33.141129 -39.668289 -12.076756
* Map size: 89 81 75
Receptor desolvation map information:
* Receptor electrostatic map read from file
* Map origin: -24.141129 -30.668287 -3.076756
* Map size: 71 63 57
* Extra potential receptor maps
*Number of read potentials: 0
Reading ligand pdb
* Ligand center: 229.958908 23.243317 49.012619
* Ligand accesilibility read from Ligand PDB.
Checking ASA in Occupancy column...
- Ok. Occupancy column seems to show ASA values
* Ligand desolvation map read from file
Spherical radius
* Receptor vdw radius in amstrongs: 71.000000 (71 spherical layers)
* Ligand radius in amstrongs: 28.025059 (29 spherical layers)
* Ligand electrostatic radius in amstrongs: 28.025059 (29 spherical layers)
* Ligand accesibility radius in amstrongs: 28.025059 (29 spherical layers)
* Ligand desolvation radius in amstrongs: 33.025059 (34 spherical layers)
* Maximum ligand desolvation radius in amstrongs: 33.025059 (34 spherical layers)
Precomputation of spherical representation of ligand
Precomputation Fixed time: 43.110000 s
Determination of translational points to explore
* Mask created
* Number of points to explore: 18996
Translational search init...
* Translational search finished
* Number of solutions: 69464
--------------- STEP 4. Clustering and visualization of predicitions ---------------
Clusterization of the solutions file is done by:
Sort process...
Maximum correlation: 1299.208740
Clustering process...
As before, the solutions can be displayed using frodockview. The closest first solution can be found at 11th position. To get the coordinates of such prediction type:
11 120.7 152.8 82.0 19.3 8.8 55.4 1161.89 => 1lza_ly2_11.pdb
You can visualize this good solution (1lza_ly2_11.pdb, cyan) with your favorite molecular viewer. Note the proximity to correct solution (3hfl_ly2_ref.pdb, red).
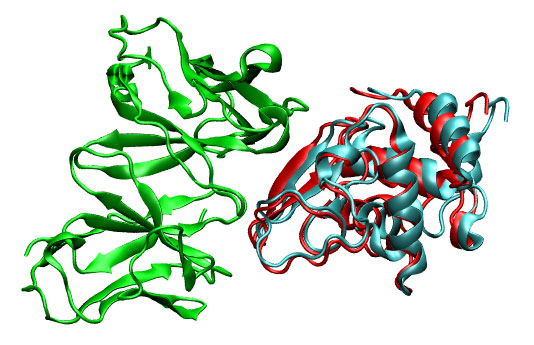 |
FRODOCK_MPI versions works in the same way as the sequential version, but using mpirun it is possible to split the traslational search in as many parallel processes as desired. To this end:
--------------- STEP 3. Perform the exhaustive docking search ---------------
This command computes the docking process using 6 parallel processes. The frodock_mpi binary files requires dynamic links to a set of MPI (version 1.3.1) and system libraries. If the corresponding frodock_mpi does not work correctly with your MPI installation, check whether the libraries are accessible by:
linux-gate.so.1 => (0xffffe000)
libmpi_cxx.so.0 => /usr/lib/openmpi/1.4-gcc/lib/libmpi_cxx.so.0(0xf7ef7000)
libmpi.so.0 => /usr/lib/openmpi/1.4-gcc/lib/libmpi.so.0 (0x00c29000)
libopen-rte.so.0 => /usr/lib/openmpi/1.4-gcc/lib/libopen-rte.so.0 (0x00b7d000)
libopen-pal.so.0 => /usr/lib/openmpi/1.4-gcc/lib/libopen-pal.so.0 (0x00b0c000)
etc...
If any of the libraries listed is marked up as "Not Found" it will be necessary to install it or include its path in LD_LIBRARY_PATH (NOTE: It is possible that existing libraries present a different name, in that case a symbolic link can solve this problem).